September 25, 2020
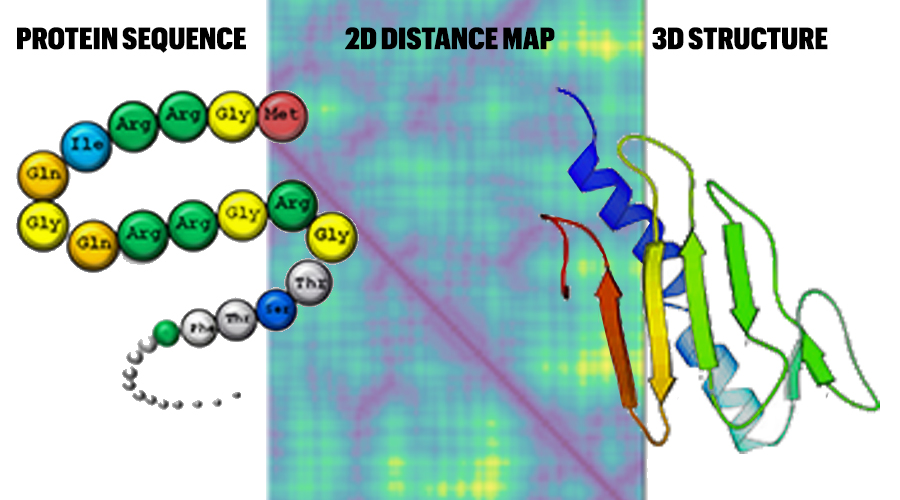
Using artificial intelligence technologies, Thompson Professor Jianlin Cheng is devising a way to predict 3-dimensional protein structures. The method translates a protein sequence into a 2-dimensional map to predict the structure.
Knowing the three-dimensional structure of proteins—such as the shape of the spike-like protein that injects coronavirus into our cells—can help us treat illnesses. That’s one reason why predicting protein structures remains one of the world’s highest health priorities.
Mizzou Engineering Jianlin Cheng has been working on protein prediction methods for more than a decade. Cheng, the Thompson professor in electrical engineering and computer science, and collaborator John Tanner, associate chair of biochemistry in the College of Agriculture, Food and Natural Resources, are devising a computational model to predict protein structures.
Earlier this month, they received a $1.37 million grant from the National Institutes of Health to further develop the prediction method.
Explainable AI Prediction
Proteins are made up of strings of amino acids that fold into specific structures. Those shapes determine what functions a protein will carry out.
Today, few protein structures are known. However, the individual amino acids that make up protein sequences have been identified.
“Our project uses computers to predict the structure from the protein’s sequence,” Cheng said. “You give the computer the sequence information and it builds the predicted shape for this sequence.”
Cheng was one of the first to use deep learning methods—a cutting-edge artificial intelligence (AI) technology—to more easily predict protein structures based on their specific amino acids and how far apart those molecules are from one another. While researchers around the world are now using deep learning to study the issue, Cheng’s model is unique in that it provides explainable information to support the prediction.
By predicting or even controlling protein structures, scientists can design drugs to develop more precise medicine.
“If we know the structure, we can find specific molecules to develop drugs,” Cheng said. “We can see which molecules interact with the protein’s structure. In the coronavirus, we can find something that interacts with some area of the spike protein and then neutralize it. That’s why the structure is very important.”